Research
We have been investigating a variety of antifungal proteins for sutability. However, due to the advantages of a short sequence, we focused on antifungal protiens with a length of less than 50 amino acids. Of these, we focused on two classes: the chitin synthesis inhibitors and the glucan synthesis inhibitors, due to the fact that fungal cell wals are made of chitin and glucan.
We found 5 promising protiens that we investigated further, using a discussion by De Lucca et al.
Chitin synthesis inhibitors:
nikkomycin Z (Li et al)
polyoxin D (Endo et al)
FR900403 (Iwamoto et al)
Glucan sythesis inhibitors:
Pneumocandin B0 (Youssar et al)
aculeacin A (Inokoshi et al)
However, none of these protiens were good candidates for transforming our E. coli bacteria. Many we could not even find the sequences for, so we searched for a new possibility.
For the second time, we did a search of the NCBI database looking for chitinases, since chitin makes up fungal cell walls. In it, we restricted ourselves to protiens that met two characteristics:
- Less than 500 amino acids in length. We wished our plasmid to be around 5000 total base pairs, as we have had success transforming that particular strain of E. coli with plasmids of that length. We had initially looked for proteins with less than 50 base pairs so that they could diffuse out of the E. coli without a seperate release mechanism. As we had had little success, we decided to go with a longer sequence and lyse the cells to release the protien, if necessary.
- Bacterial in origin. We did not want a protein that would kill our E. coli chassis, so we selected a bacterial one to make sure that it was not deadly to bacteria
We came up with one protein that fulfilled all of our requirements: Chitinase D, a 488 amino acid protien from Bacillus circulans that attacked N-acetyl-beta-D-glucosaminide (1->4)-beta-linkages in chitin. It had a gene sequence, too. ( Watanabe et al) ( UNIPROT databse)
For our plasmid, we wanted three things:
- A pre-synthesized and well-documented part, to avoid wasting time and money synthesizing it ourselves.
- High copy number, as these produce more protien. We looked at medium-low and low copy plasmids too, however the benefit they offered, reduced use of cellular resourced leading to a more consistent performace, would be less of a concern to us, as we are not running multiple plasmids in parallel, and production inconsistencies would not be a problem.
- We preferred to have ampicillin resistance, as this is a very standard resistance that we have had experience in using.
Looking through the possible plasmids on parts.igem.org, we came up with two candidates:
- pSB1C3, a 2070 bp plasmid which came with a high-copy origin of replication, chloroamphenicol resistance, promoter, terminator. This is the iGEM registry standard plasmid.
- pSB1A3, a 2155 bp plasmid that was very similar to the first one, except that it had ampicillin resistance. There were also teams that had reported it to work. It has several problems, such as having nonfunctional EcoR1 and Pst1 sites and being difficult to turn off, but as our project only requires expression of one protien, these issues did not seem important.
We ended up deciding that pSB1C3 was the better choice, as it being the iGEM registry standard protein would ensure that it would be functional for a wide range of proteins.
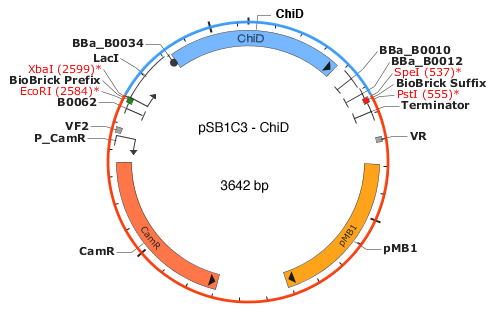
A map of the plasmid. With our gene, the total length would come to 3642 base pairs, so it would be a good length to transform. (Map adapted from http://parts.igem.org/wiki/index.php/Part:pSB1C3)
