Labs

What a Colorful World:
Purpose:
Conduct and interpret the results of a bacterial transformation
Procedure
Part 1: Preparing Strain 4-1 and 4-2 for transformation
Neither of these E. coli strains will take up DNA from the environment until they are treated with a salt solution that makes their outer membrane slightly porous. The cells will become "competent" for transformation (i.e. ready to bring DNA that's external to the cell into the cytoplasm where the DNA code can be expressed). The cells will also become fragile. Keep the cells cold and don't pipet them roughly once you have swirled them into the CaCl2 salt solution.
- Label 2 microcentrifuge tubes either "4-1" for E. coli. strain 4-1 or "4-2" for E. coli. strain 4-2.
- Pipet 200 ul of CaCl2 solution into each tube and then place the tubes on ice.
- Use a sterile wooden dowel to scrape up one entire patch of cells (NOT including the agar that they're growing on!) labeled "4-1," and then swirl the cells into its tube of cold CaCl2. Gently flick and invert the tube, then return it to your ice bucket.
- Repeat, using a different sterile wooden dowel to scrape up the patch of cells labeled "4-2.". It's OK for some clumps of cells to remain in this solution.
- Keep these competent cells on ice while you prepare the DNA for transformation.
Part 2: Transforming Strains 4-1 and 4-2 with pPRL and pGRN

You will transform the purple-color generator into each strain, and also the green-color generator into each strain. You will also use the last bit of competent cells as negative controls for the transformation.
- Retrieve 2 aliquots of each plasmid for a total of 4 samples (2x pPRL, 2x pGRN). Each aliquot has 5 ul of DNA in it. The DNA is at a concentration of 0.04 ug/ul. You will need these values when you calculate the transformation efficiency at the end of this experiment.
- Label one of the pPRL tubes "4-1." Label the other pPRL tube "4-2." Be sure that the labels are readable. Place the tubes in the ice bucket.
- Label one of the pGRN tubes "4-1." Label the other pGRN tube "4-2." Be sure that the labels are readable. Place the tubes in the ice bucket.
- Flick the tube with the competent 4-1 strain and then pipet 75 ul of the bacteria into the tube labeled "pPRL, 4-1" and an additional 75 ul into the tube labeled "pGRN, 4-1." Flick to mix the tubes and return them to the ice. Save the remaining small volume of the 4-1 strain on ice.
- Flick the tube with the competent 4-2 strain and then pipet 75 ul into the tube labeled "pPRL, 4-2" and an additional 75 ul into the tube labeled "pGRN, 4-2." Flick to mix and store them, as well as the remaining volume of competent cells, on ice.
- Let the DNA and the cells sit on ice for 5 minutes.
- While your DNA and cells are incubating, you can label the bottoms (not the tops) of the 6 petri dishes you'll need. The label should indicate the strain you've used ("4-1" or "4-2") and the DNA you've transformed them with ("pPRL" and "pGRN").
- Heat shock all of your DNA/cell samples by placing the tubes at 42 degrees for 90 seconds exactly (use a timer). This step helps drive the DNA into the cells and closes the porous bacterial membranes of the bacteria.
- At the end of the 90 seconds, move the tubes to a rack at room temperature.
- Add 0.5 ml of room temperature LB to the tubes. Close the caps, and invert the tubes to mix the contents.
- Using a sterilized spreader or sterile beads, spread 250 ul of the transformation mixes onto the surface of LB+ampicillin agar petri dishes.
- If desired the remaining volumes of transformation mixes can be plated on LB plates to show the effect of antibiotic selection on the outcome.
- Incubate the petri dishes with the agar side up at 37° overnight, not more than 24 hours.
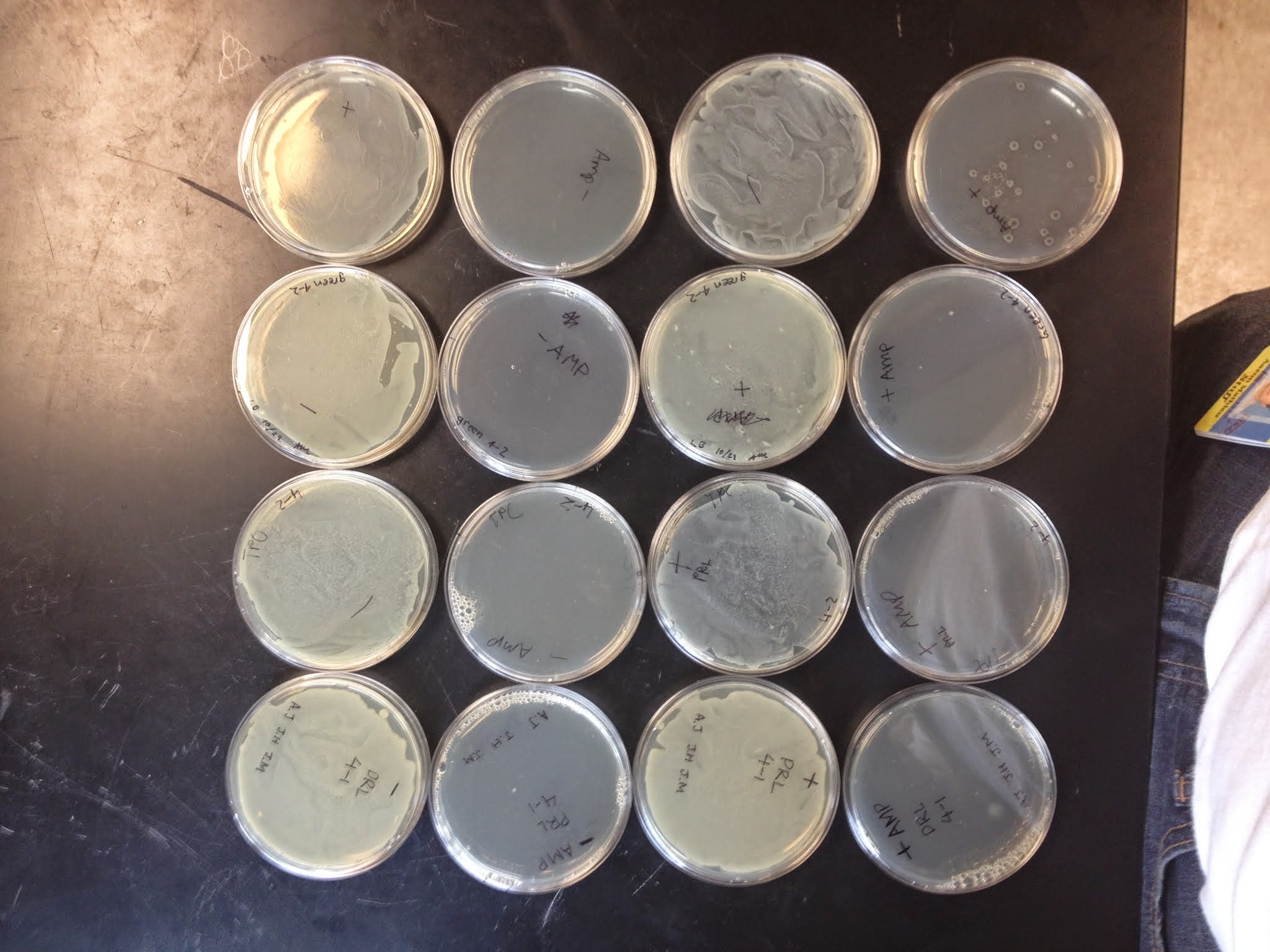
Part 3: Collecting the Data
- Count the number of colonies growing on each petri dish.
- Small white colonies that are growing around the perimeter of larger colored colonies are called "satellites." They should not be counted. They grow near the central colony only after the cells there have inactivated the ampicillin that's in the petri dish agar.
- You can feel most confident in your results if there are between 20 and 200 colonies on the petri dish. Fewer than 20 and your value is affected by errors in pipeting that make large percentage differences in the outcome. Greater than 200 colonies and they become hard to count reliably. If the petri dish has many colonies growing on it, try to divide the dish into pie sections (1/4th or 1/8ths or even 1/16ths of the area), and then count a representative area. Finally, multiply the number you get for the section to get your total number of colonies.
- Based on the number of colonies you find on each petri dish, calculate the transformation efficiency for each. Transformation efficiency is a measure for how well the cells incorporated the DNA. The units for transformation efficiency are "colonies per microgram of DNA." Each transformation used 200 nanograms (=0.2 micrograms) of DNA and you plated only 1/2 the transformation mixes on the petri dishes.
- Record the color of the colonies you see.
